
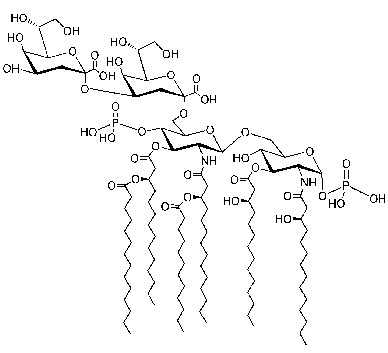
Kdo2 Lipid A (E. coli-type Re LPS)
Lipopolysaccharide (LPS) is an amphiphilic glucosamine-based phospholipid found on the outer leaflet of outer membranes of most Gram-negative bacteria. LPS activates the immune system in mammals and is considered to play a key role in human septic shock syndrome. Recent studies on the mechanism of LPS-induced immunostimulation have led to great progress in our understanding of protein linkage in the signaling pathway. The answer to a basic question remains unclear, however: Is the active unit of LPS mono- or multimeric. To answer this question, extensive studies on the transition of LPS from monomers to micelles have been needed.
Some groups have reported on studies of the aggregation behavior using fluorescence spectroscopy, light scattering, and equilibrium dialysis. Data obtained by fluorescence or light scattering, however, did not actually show the monomer-micelle transition, due to the sensitivity limit of the applied measuring techniques. Moreover, the results from equilibrium dialysis indicated only the maximum concentration at which LPS could exist as monomers (e.g., E. coli-type deep rough mutant (Re) LPS: 33 nM at 22°C).
Using a chemically synthesized pure E. coli-type Re LPS, which was specially prepared as Kdo2 Lipid A by Avanti Polar Lipids, Inc. for the LIPID MAPS project, we have investigated the transition from monomers to micelles as a function of LPS concentration, using high-sensitivity dynamic light scattering and densitometry.
Instrument: Wyatt DAWN HELEOS utilizing 690 nm laser at 25°C
Sample: Prepared by diluting the extruded lipid solution with DPBS.
Cumulants method analysis:
![]()
Γ: Mean decay rate
μ: Variance of the distribution of Γ
Rh = KΒΤq2/(6πηΓ)
Rh: Hydrodynamic radius
Frisken, B. J. Appl. Optics 2001, 40, 4087.
Instrument: DMA 10 digital precision density meter at 25°C. Density of solution (dsolution) was determined from the variance in the natural frequencies of a hollow oscillator filled with sample and water.
Sample: Kdo2 Lipid A (0.07450 ± 0.00003 g) in water (1.52255 ± 0.00093 g)
Calculation of molecular volume of Kdo2 lipid A (VL):
dL = WL / [(Wwater + WL) / dsolution - Wwater / dwater]
dL, dwater, dsolution : Densities of the lipid, water and solution
WL, Wwater : Weights of the lipid and water
VL = Mw / (dLNA)
Mw : Molecular weight of the lipid
NA : Avogadro's number
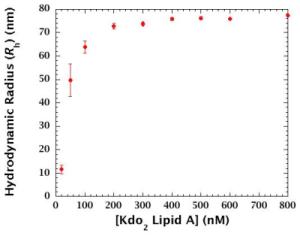
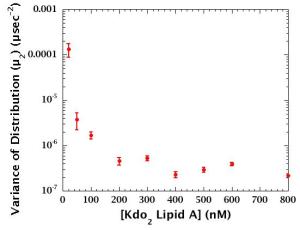
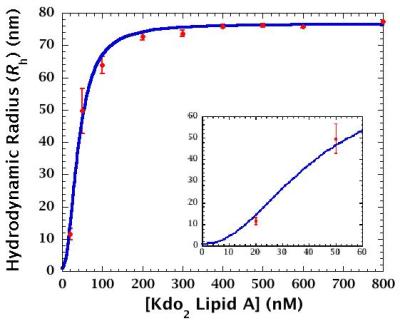
> 200 nM: Aggregates are predominant component.
< 200 nM: Monomer-to-aggregate ratio and/or aggregate size changes.
|
|
Density of Kdo2 Lipid A (dL) = 1.21266 ± 0.02719 (g/cm3)
Molecular volume of Kdo2 Lipid A (VL) = 3158.73 ± 70.82 (Å3)
=> Estimated radius of Kdo2 Lipid A monomer = 0.91 (nm)
Gradual aggregation occurs in the range of 2-200 nM.
This work was supported by the LIPID MAPS Large-Scale Collaborative Grant GM-069338 from the National Institute of Health.